Chemical reactions¶
In the previous tutorial Defining a model, we defined a microbenthic domain and a single variable to construct a single differential equation. We will now create chemical reactions between two solutes and study the dynamics and distributions of their variables by solving couple partial differential equations.
See the definition file for this tutorial.
Firstly, we define another diffusive solute hydrogen sulfide \(H_2S\) under environment.
49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 | h2s:
cls: ModelVariable
init_params:
name: h2s
create:
hasOld: true
value: !unit 0.0 mol/m**3
constraints:
top: !unit 10.0 mumol/l
bottom: !unit 1e-3 mol/l
clip_min: 0.0
seed:
profile: linear
D_h2s:
cls: Process
init_params:
expr:
formula: porosity * D0_h2s
params:
D0_h2s: !unit 0.02 cm**2/h
|
Respiration¶
We will now specify reactions that involve the oxy and h2s variables, which will be cast
as “source” terms in the differential equations. First we specify that the oxygen within the
sediment is consumed through aerobic respiration in the environment.
76 77 78 79 80 81 82 83 | aero_respire:
cls: Process
init_params:
expr:
formula: -Vmax * porosity * sed_mask * saturation(oxy, Km)
params:
Vmax: !unit 1.0 mmol/l/h
Km: &aero_Km !unit 1e-5 mol/l
|
This specification states that the respiration process at a rate of Vmax. Since respiration
does not occur within the sediment grains but within the porespaces, we multiply it by
porosity, which is defined from the model domain. We want that the reaction occurs only in
the sediment and not in the water column, so we use the variable sed_mask. This
merely selects the region of the domain that is the sediment. Additionally, we specify that the
rate of respiration has a saturation dependence on oxygen, that is the rate of the process slows
down at high enough levels (parameterized by Km) of oxygen. What is the formulation for the
saturation(oxy, Km) function? It can be specified under the namespace key of the process’
init_params. Alternatively, if a formula is to be reused then in a formulae
section of the model as follows.
121 122 123 124 125 | formulae:
saturation:
vars: [x, Km]
expr: x / (Km + x)
|
Abiotic sulfide oxidation¶
Another process that occurs in sedimentary system is the abiotic oxidation of sulfide. That is
oxygen reacts with hydrogen sulfide in a 2:1 stoichiometry. We can define this process also in
the environment.
87 88 89 90 91 92 93 | abio_sulfoxid:
cls: Process
init_params:
expr:
formula: porosity * k * oxy * oxy * h2s * sed_mask
params:
k: !unit -70.0 1/h/(mmol/l)**2
|
This reaction process therefore couples the equations of the two variables oxy and h2s,
that so far had no shared process terms. The definition of this process should therefore appear
in both equations.
The equations for this model will therefore be:
97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 | equations:
oxyEqn:
transient: [domain.oxy, 1]
diffusion: [env.D_oxy, 1]
sources:
- [env.abio_sulfoxid, 2]
- [env.aero_respire, 1]
h2sEqn:
transient: [domain.h2s, 1]
diffusion: [env.D_h2s, 1]
sources:
- [env.abio_sulfoxid, 1]
|
Note that the stoichiometry of the sulfide oxidation process is represented by a coefficient of 2
in the oxygen equation, indicating that for each H2S consumed two O2 are consumed by this
process. The definition of the environmental process abio_sulfoxid enables us to easily
represent the process in multiple equations.
Run it¶
This creates the equation to solve
Running the model simulation with:
microbenthos -v simulate definition_input.yml --plot --show-eqns
should show the equation in the console and open up a graphical view of the model as it is simulated.
An extracted frame is shown below.
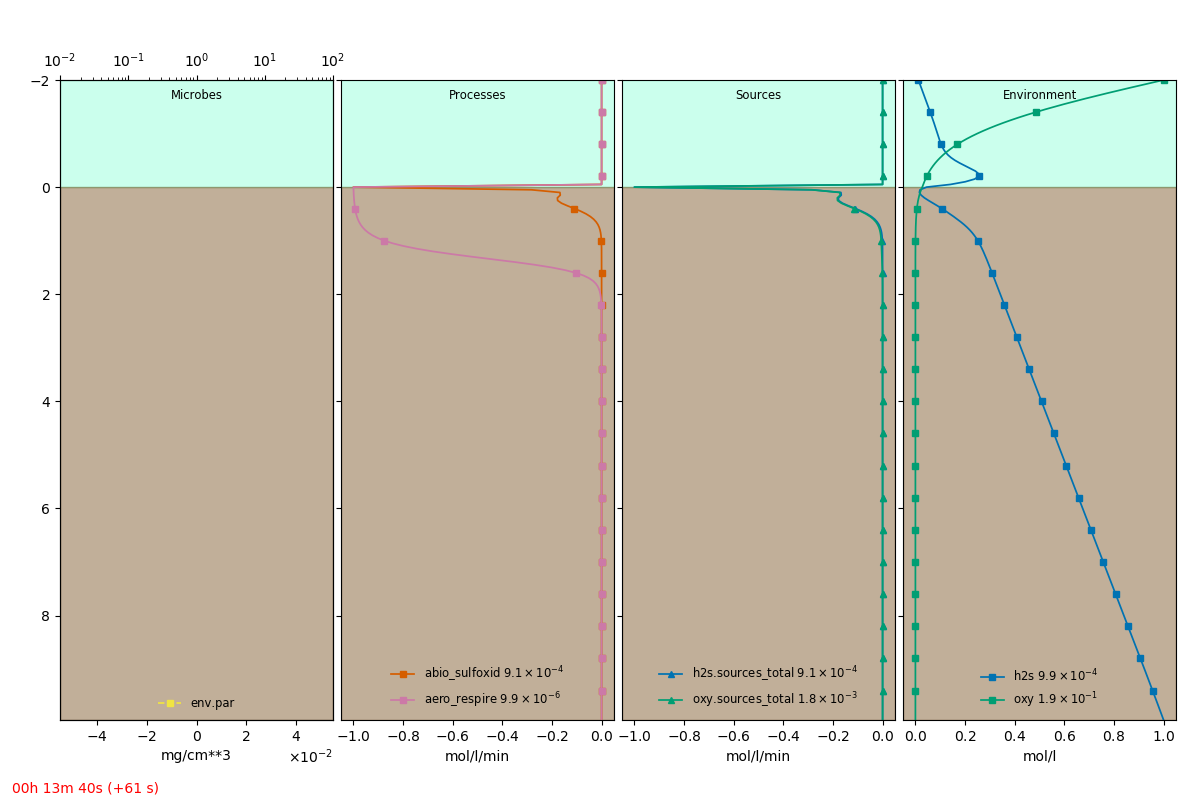
The full definition file is:
model:
domain:
cls: SedimentDBLDomain
init_params:
cell_size: !unit 50 mum
sediment_length: !unit 10 mm
dbl_length: !unit 2 mm
porosity: 0.6
environment:
irradiance:
cls: Irradiance
init_params:
hours_total: !unit 4h
day_fraction: 0.5
channels:
- name: par
k0: !unit 15.3 1/cm
oxy:
cls: ModelVariable
init_params:
name: oxy
create:
hasOld: true
value: !unit 0.0 mol/m**3
constraints:
top: !unit 230 mumol/l
bottom: !unit 0.0 mol/l
seed:
profile: linear
D_oxy:
cls: Process
init_params:
expr:
formula: porosity * D0_oxy
params:
D0_oxy: !unit 0.03 cm**2/h
# start: h2s
h2s:
cls: ModelVariable
init_params:
name: h2s
create:
hasOld: true
value: !unit 0.0 mol/m**3
constraints:
top: !unit 10.0 mumol/l
bottom: !unit 1e-3 mol/l
clip_min: 0.0
seed:
profile: linear
D_h2s:
cls: Process
init_params:
expr:
formula: porosity * D0_h2s
params:
D0_h2s: !unit 0.02 cm**2/h
# stop: h2s
# start: aero_respire
aero_respire:
cls: Process
init_params:
expr:
formula: -Vmax * porosity * sed_mask * saturation(oxy, Km)
params:
Vmax: !unit 1.0 mmol/l/h
Km: &aero_Km !unit 1e-5 mol/l
# stop: aero_respire
# start: abio_sulfoxid
abio_sulfoxid:
cls: Process
init_params:
expr:
formula: porosity * k * oxy * oxy * h2s * sed_mask
params:
k: !unit -70.0 1/h/(mmol/l)**2
# stop: abio_sulfoxid
# start: equations
equations:
oxyEqn:
transient: [domain.oxy, 1]
diffusion: [env.D_oxy, 1]
sources:
- [env.abio_sulfoxid, 2]
- [env.aero_respire, 1]
h2sEqn:
transient: [domain.h2s, 1]
diffusion: [env.D_h2s, 1]
sources:
- [env.abio_sulfoxid, 1]
# stop: equations
# start: formulae
formulae:
saturation:
vars: [x, Km]
expr: x / (Km + x)
# stop: formulae
simulation:
simtime_total: !unit 8h
simtime_lims: [0.1, 180]
max_residual: 1e-13