Microbial groups¶
In the previous tutorial Chemical reactions, we defined a microbenthic domain with two reacting chemical solutes in the environment. We will now define specific microbial populations or groups which perform specific metabolic processes involving the solutes. This tutorial derives conceptually from de Wit et al (1995), but modifies it to focus on metabolic rates instead of biomass growth. The formulations of the constitutive relationships between metabolisms and environmental parameters are used from the paper, and the tutorial mainly indicates how these symbolic relationships can be used to construct and solve coupled partial differential equations.
See the definition file for this tutorial.
See also
Rutger de Wit, Frank P. van den Ende, Hans van Gemerden; Mathematical simulation of the interactions among cyanobacteria, purple sulfur bacteria and chemotrophic sulfur bacteria in microbial mat communities, FEMS Microbiology Ecology, Volume 17, Issue 2, 1 June 1995, Pages 117–135 (DOI)
Cyanobacteria: biomass¶
We will define a distribution of cyanobacterial population in the microbenthic domain, which will perform
respiration
oxygenic photosynthesis
sulfide-driven anoxygenic photosynthesis
Microbial groups are defined under the microbes key under model. Microbial groups contain
features, which represent variables related to the population, the primary one being biomass.
The definition of the cyano group could be specified as:
95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 | microbes:
cyano:
init_params:
name: cyano
features:
biomass:
init_params:
name: biomass
create:
value: !unit 0 kg/m**3
seed:
profile: normal
params:
loc: !unit 1 mm
scale: !unit 2 mm
coeff: !unit 12 mg/cm**3
|
We have specified a (typically required) feature called biomass that has the units of
\(kg/m^3\), and is seeded with a normal distribution centered at 1 mm depth with a width of 2
mm, reaching a maximum value of \(12 mg/cm^3\).
Light absorption¶
Phototrophs, such as cyanobacteria, absorb light energy to use in their photosynthetic reactions.
The biomass-related depletion of the irradiance can be specified by modifying the irradiance
channel par as follows.
21 22 23 24 25 | channels:
- name: par
k0: !unit 15.3 1/cm
k_mods:
- [microbes.cyano.biomass, !unit 7687.5 cm**2/g]
|
This specifies that an additional attenuation based on the biomass should occur in the photosynthetically active radiation.
Oxygenic photosynthesis¶
Cyanobacteria perform oxygenic photosynthesis, i.e. they use the absorbed light energy to fix
carbon into their biomass and release dioxygen as a side product. For the current case, we ignore
any changes in biomass and consider the model dynamics for a short period of 4 hours. Oxygenic
photosynthesis produces oxygen, and the rate of this production would depend on the available
light and biomass at a specific depth in the microbial mat. So approximately, we consider the
oxygenic photosynthesis rate as \(Qmax \cdot biomass \cdot optimum(par, Ks, Ki)\), where the
optimum function (unitless) describes the irradiance intensities which are optimal for the
biomass. However, it is known that oxygenic photosynthesis is inhibited by the presence of
sulfide and oxygen itself. These are represented through an inhibition function depending on a
particular variable. So the formulae we need are:
201 202 203 204 205 206 207 208 209 210 211 212 213 | formulae:
saturation:
vars: [x, Km]
expr: x / (Km + x)
optimum:
vars: [x, Ks, Ki]
expr: x/(x + Ks)/(1 + x/Ki)
inhibition:
vars: [x, Kmax, Khalf]
expr: (Kmax - x) / (2*Kmax - Khalf - x) * (x < Kmax)
|
Using these in the model/formulae section, we can now define the oxygenic photosynthesis
process in the cyano group as:
113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 | processes:
oxyPS:
cls: Process
init_params:
params:
Ks: 1
Ki: 10
Qmax: !unit 8.4 mmol/g/h
Kmax: !unit 0.35 mmol/l
Khalf: !unit 0.3 mmol/l
Kmax2: !unit 0.8 mmol/l
Khalf2: !unit 0.7 mmol/l
expr:
formula: "Qmax * biomass * sed_mask * optimum(par, Ks, Ki)
* inhibition(h2s, Kmax, Khalf)
* inhibition(oxy, Kmax2, Khalf2)"
implicit: false
|
Note here that the biomass variable is sourced from the cyano microbial group, which
behaves as the domain. Variables not found in the group are then looked up from the group
containing the cyano group, that is the model. So the model domain is used to source the
par and h2s variables.
This formulation is straightforward to write, but pops up a mathematical problem. The inhibition
function contains a \(x < Kmax\) term, which makes the function only piecewise polynomial.
Piecewise functions cannot be generally considered differentiable. This affects the casting of
the process expression as an implicit source term (see fipy docs about implicit terms), since
it tries to estimate the first derivative of the function with respect to the variable. So here
we can simply turn off this behavior on a per-process level, by setting implicit: false in
the initialization parameters.
However, for complex non-linear source terms it is advantageous to be able to specify the pieces of the piece-wise response in the equation terms. MicroBenthos allows you to therefore specify the inhibition response of oxygen to the oxygen concentration itself by the formulation below.
135 136 137 138 139 140 141 142 | expr:
formula:
base: "Qmax * biomass * sed_mask * optimum(par, Ks, Ki)
* inhibition(h2s, Kmax, Khalf)"
pieces:
- expr: (Kmax2 - oxy) / (2*Kmax2 - Khalf2 - oxy)
where: oxy < Kmax2
|
In this formulation, we replaced the inhibition(oxy) term from before, and defined a
piecewise response of the process ourselves. The formulation of inhibition is taken as
before, but without the ( x < Kmax ) term, and is instead set as a piece mask in where.
Now, we do not need to set implicit: false, as each of the defined pieces in expr is
differentiable and MicroBenthos will handle the symbolic math accordingly to cast the overall
expression as an implicit source, if required.
Anoxygenic photosynthesis¶
Cyanobacteria also can use other electron donors than water, specifically, they can drive carbon
fixation through sulfide-driven photosynthesis. This process uses \(H_2S\) instead of
\(H_2O\). We consider that this process is also driven by \(Qmax \cdot biomass \cdot
optimum(par, Ks, Ki)\), and further modulated by metabolic response functions. We consider that
the cyanobacterial population responds to the sulfide as an optimum function. So we write the
process for anoxygenic photosynthesis as follows.
146 147 148 149 150 151 152 153 154 155 156 157 158 159 | anoxyPS:
cls: Process
init_params:
expr:
formula: "Qmax * biomass * sed_mask * optimum(par, Ks, Ki)
* optimum(h2s, Ksh2s, Kih2s)"
params:
Ks: 1
Ki: 10
Qmax: !unit -1.2 mmol/g/h
Ksh2s: !unit 900 mumol/l
Kih2s: !unit 3 mmol/l
|
Respiration¶
We can also include biomass-dependent respiration by the cyanobacteria itself. Similar to the sedimentary aerobic respiration case, the process rate saturates with respect to oxygen.
163 164 165 166 167 168 169 170 | respire:
cls: Process
init_params:
expr:
formula: Qmax * biomass * sed_mask * saturation(oxy, Km)
params:
Qmax: !unit -0.0002 mumol/g/h
Km: !unit 1e-6 mol/l
|
We note in all the cases above, that each process essentially provides a local “namespace” to avoid clash of identically named parameters in other processes. Additionally, the microbial group provides a local store of variables, like the model domain, and passes forward the look up for missing variables to the model domain itself.
Run it¶
This creates the equations to solve
Running the model simulation with:
microbenthos -v simulate definition_input.yml --plot --show-eqns
should show the equation in the console and open up a graphical view of the model as it is simulated.
An extracted frame is shown below.
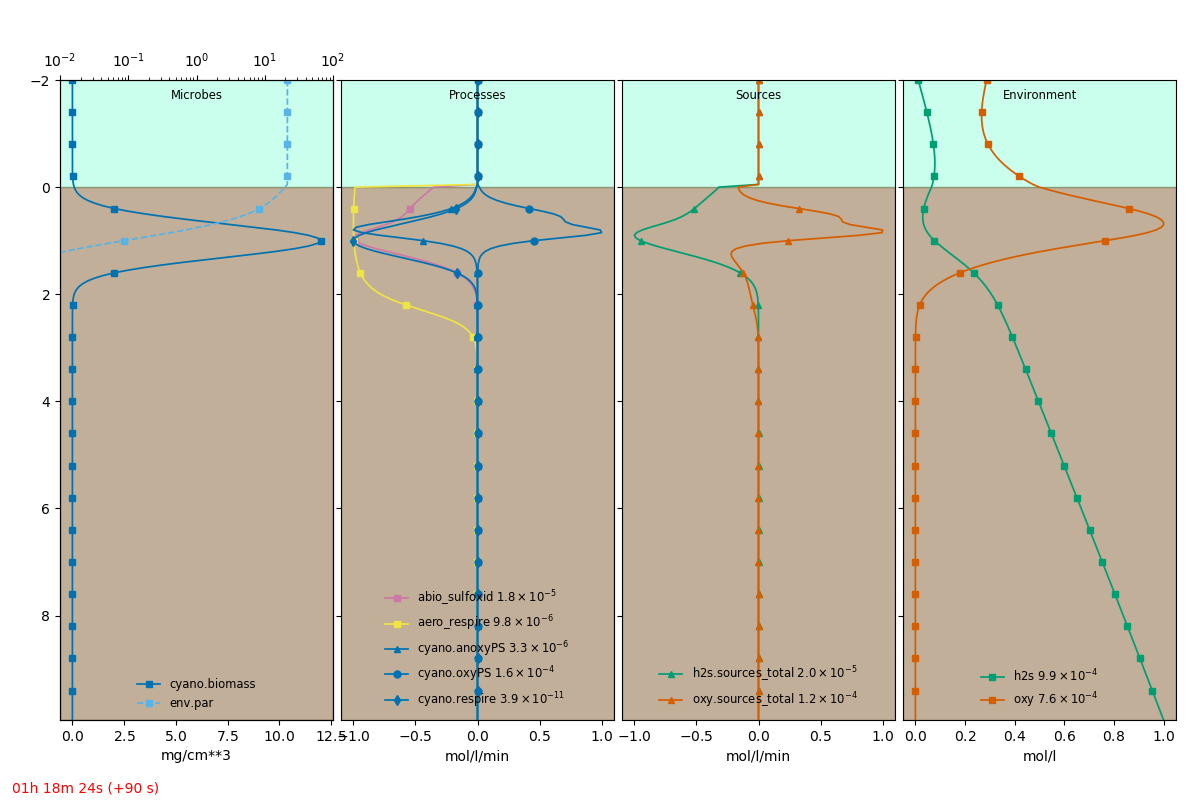
The full definition file is:
model:
domain:
cls: SedimentDBLDomain
init_params:
cell_size: !unit 50 mum
sediment_length: !unit 10 mm
dbl_length: !unit 2 mm
porosity: 0.6
environment:
irradiance:
cls: Irradiance
init_params:
hours_total: !unit 4h
day_fraction: 0.5
# start: channels
channels:
- name: par
k0: !unit 15.3 1/cm
k_mods:
- [microbes.cyano.biomass, !unit 7687.5 cm**2/g]
# stop: channels
oxy:
cls: ModelVariable
init_params:
name: oxy
create:
hasOld: true
value: !unit 0.0 mol/m**3
constraints:
top: !unit 230.0 mumol/l
bottom: !unit 0.0 mol/l
seed:
profile: linear
D_oxy:
cls: Process
init_params:
expr:
formula: porosity * D0_oxy
params:
D0_oxy: !unit 0.03 cm**2/h
h2s:
cls: ModelVariable
init_params:
name: h2s
create:
hasOld: true
value: !unit 0.0 mol/m**3
constraints:
top: !unit 10.0 mumol/l
bottom: !unit 1e-3 mol/l
seed:
profile: linear
clip_min: 0.0
D_h2s:
cls: Process
init_params:
expr:
formula: porosity * D0_h2s
params:
D0_h2s: !unit 0.02 cm**2/h
aero_respire:
cls: Process
init_params:
expr:
formula: -Vmax * porosity * sed_mask * saturation(oxy, Km)
params:
Vmax: !unit 1.0 mmol/l/h
Km: &aero_Km !unit 1e-5 mol/l
abio_sulfoxid:
cls: Process
init_params:
expr:
formula: porosity * sed_mask * k * oxy * oxy * h2s
params:
k: !unit -70.0 1/h/(mmol/l)**2
# start: microbes
microbes:
cyano:
init_params:
name: cyano
features:
biomass:
init_params:
name: biomass
create:
value: !unit 0 kg/m**3
seed:
profile: normal
params:
loc: !unit 1 mm
scale: !unit 2 mm
coeff: !unit 12 mg/cm**3
# stop: microbes
# start: oxyPS1
processes:
oxyPS:
cls: Process
init_params:
params:
Ks: 1
Ki: 10
Qmax: !unit 8.4 mmol/g/h
Kmax: !unit 0.35 mmol/l
Khalf: !unit 0.3 mmol/l
Kmax2: !unit 0.8 mmol/l
Khalf2: !unit 0.7 mmol/l
expr:
formula: "Qmax * biomass * sed_mask * optimum(par, Ks, Ki)
* inhibition(h2s, Kmax, Khalf)
* inhibition(oxy, Kmax2, Khalf2)"
implicit: false
# stop: oxyPS1
# start: oxyPS2
expr:
formula:
base: "Qmax * biomass * sed_mask * optimum(par, Ks, Ki)
* inhibition(h2s, Kmax, Khalf)"
pieces:
- expr: (Kmax2 - oxy) / (2*Kmax2 - Khalf2 - oxy)
where: oxy < Kmax2
# stop: oxyPS2
# start: anoxyPS
anoxyPS:
cls: Process
init_params:
expr:
formula: "Qmax * biomass * sed_mask * optimum(par, Ks, Ki)
* optimum(h2s, Ksh2s, Kih2s)"
params:
Ks: 1
Ki: 10
Qmax: !unit -1.2 mmol/g/h
Ksh2s: !unit 900 mumol/l
Kih2s: !unit 3 mmol/l
# stop: anoxyPS
# start: respire
respire:
cls: Process
init_params:
expr:
formula: Qmax * biomass * sed_mask * saturation(oxy, Km)
params:
Qmax: !unit -0.0002 mumol/g/h
Km: !unit 1e-6 mol/l
# stop: respire
equations:
oxyEqn:
transient: [domain.oxy, 1]
diffusion: [env.D_oxy, 1]
sources:
- [env.abio_sulfoxid, 2]
- [env.aero_respire, 1]
- [microbes.cyano.processes.oxyPS, 1]
- [microbes.cyano.processes.respire, 1]
h2sEqn:
transient: [domain.h2s, 1]
diffusion: [env.D_h2s, 1]
sources:
- [env.abio_sulfoxid, 1]
- [microbes.cyano.processes.anoxyPS, 1]
# start: formulae
formulae:
saturation:
vars: [x, Km]
expr: x / (Km + x)
optimum:
vars: [x, Ks, Ki]
expr: x/(x + Ks)/(1 + x/Ki)
inhibition:
vars: [x, Kmax, Khalf]
expr: (Kmax - x) / (2*Kmax - Khalf - x) * (x < Kmax)
# stop: formulae
simulation:
simtime_total: !unit 8h
simtime_lims: [0.1, 180]
max_residual: 1e-13